Introduction
According to the National Institutes of Health Biomarkers Definitions Working Group, a biomarker is defined as ‘a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention’.1 Biomarkers are employed for diagnosis, staging of disease, prognosis and monitoring the response to intervention.2 Their major limitation as diagnostic parameter is the need for evaluating and re-evaluating their validity. Some criteria for ideal tumor marker include easy and inexpensive to measure, specific to the associated tumor, stable plasma, and urine levels, preceding and predicting recurrences, etc.3
Materials and Methods
Data sources
A systematic electronic literature search was conducted to identify relevant articles from the databases of databases of PubMed/Medline and Google Scholar. The search was conducted from September 2022 to December 2022.
Search strategy
The keywords used were oral cancer, cancer, salivary, and biomarkers. They were combined with AND Boolean to generate the search syntax. The search was conducted for articles published in 2000 and onwards.
Screening and study selection
A total of 100 articles were generated following the search. After screening titles and abstracts, 30 articles were chosen for full text screening. Out of those 30, duplicate articles and articles with unobtainable access were excluded, and only 18 articles were selected based on higher academic relevance to our systematic review.
Discussion
Saliva as a source for diagnostic biomarkers
Saliva is a body fluid that is readily available and easy to collect unlike other fluids such as blood or CSF. The past concerns regarding the use of saliva for diagnosis were raised due to its low concentration of analytes in comparison to blood were dismissed by the introduction of molecular methods and nanotechnology that are highly sensitive and specific.4 The low concentration of analytes subjugated by the local origin of these biomarkers from the tumor site.5 Advantages of saliva as a diagnostic method include:4
Non-invasive
Better patient cooperation
No needle prick needed and thus, a good choice in patients who are hemophiliacs or are fearful of needles
No special equipment required for the collection and storage of saliva as saliva does not clot
Minimal risk of cross-contamination
Economical
Can be collected without trained medical personnel
Commercial availability of screening assays
As saliva has biomolecules that can inhibit HIV, the chances of transmission are lower as compared to blood samples.5
The diversity of biomarkers found in the saliva include: 6
Collection of saliva sample
The collection of saliva sample can be unstimulated or stimulated. The salivary flow rate of greater than 0.1mL/min is considered physiological for unstimulated saliva, whereas a rate of 0.2mL/min is considered for stimulated saliva.6
For the collection of the sample, the age, gender, and mouth position should be attended to while collecting the sample.7 A standardized protocol for the time of collection and amount of collection should also be established to minimize discrepancy.7
The methods for collection of saliva sample can be done following methods:7
A single protocol for saliva collection should be established to minimize the discrepancies that make occur as result of collection method such as production of stimulated saliva as a result of swabbing method or the variation in concentration of analytes.8 Collection through passive drooling has been proved to provide optimal results.9 Various commercial collection kits are available; such as Ora Sure (OraSure Technologies, Pennsylvania, USA), Salivette (Sarstedt, Germany), Salimetrics Oral Swab (SOS, California, USA), Quantisal (Abbott Rapid Diagnostics, Virginia, USA) etc.10 After collection the saliva sample, it is recommended to store the samples at -20°C or in absence of freezer at 4°C to prevent bacterial growth and degradation of salivary molecules.11
Salivary Genomic Biomarkers
The source of genetic material in saliva is from the epithelial cells that are regularly shed from the superficial layer of mucosal epithelium every 2.7 hours,12 microbial DNA13 and directly from the tumor site.5 The mean total DNA in saliva is 24 µg (range of 0.2-52 µg,). Despite the value is 10 time lower than that of blood, genotyping requires DNA material as low as 5ng/mL to work effectively.13 The pathology of tumorigenesis involves both genetic and epigenetic changes.13 The first epigenetic change to be associated with carcinogenesis is aberrant DNA methylation as a result of alteration in the normal DNA regulation.14
Study of salivary transcriptomics, as stated by Khurshid et al, assumes that salivary mRNA is enclosed in apoptotic bodies an released out in exosomes or macrovesicles actively.10 The RNAs found in saliva are produced either locally or from the serum. The methods for transportation of serum derived RNA include via acinar cells, gingiva crevicular fluid, transcellular (active transport or passive diffusion) and paracellular routes (ultrafiltration).15 Panta et al also state that in OSCC patients, the transcription of several mRNA and miRNA are altered.15 Li et al classified the genes into three groups: high upregulation mRNA (IL-8, IL-1β), moderately upregulated (S100P, H3F3A) and low upregulated mRNA (OAZ1, DUSP1).16
Biomolecular Methods for Detection of Salivary Nucleic Acids
Polymerase chain reaction (PCR)
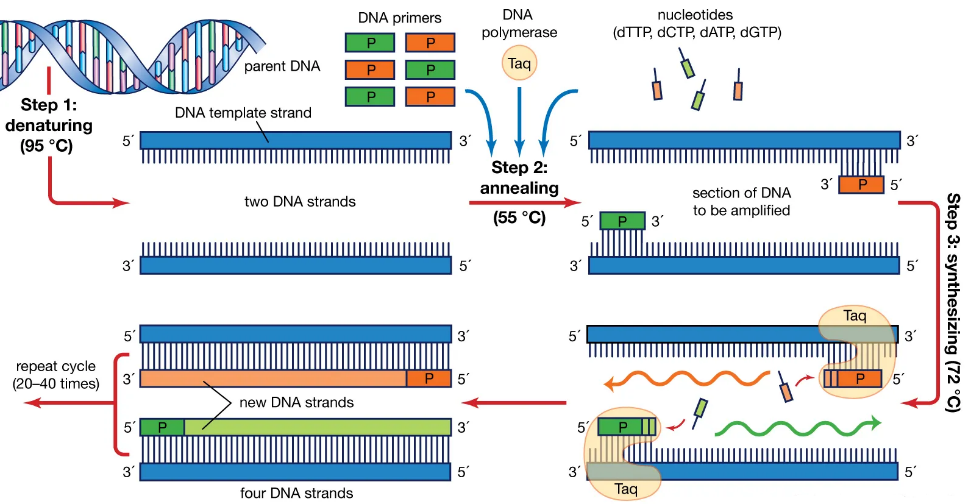
It is a simple method of amplifying short stretches of RNA or DNA sequences a few hundred base pairs long. This method uses a DNA polymerase enzyme derived from Thermus aquaticus and is commonly known as Taq DNA polymerase. It allows polymerase activity to continue after repeated cycles of denaturation and renaturation in the presence of small terminal oligonucleotides called primers. A small amount of RNA or DNA becomes quantitatively the major product by this procedure.17
There are 3 steps involved17
Denaturation: There is heating of the DNA mixture at 95OC to separate the two complementary base pairs from each other.
Annealing: The reaction mixture is cooled to between 37OC and 72OC at which the primers (20 to 30 bases long) bind to their complementary sites on the single stranded DNA molecule.
Extension/Synthetization: The temperature is raised to 72OC and the Taq polymerase enzyme begins to begins to synthesize new DNA directed from 5’ to 3’ from the priming region.
At the end of each cycle, the newly synthesized extension product serves as a template for subsequent reaction resulting in more DNA copies.17 Figure 1 depicts the above-mentioned steps that occur in PCR.
Limitations of PCR include high sensitivity for the detection of possible aerosol contamination and false-negative results due to the presence of inhibitors or poor preparation of starting materials.17
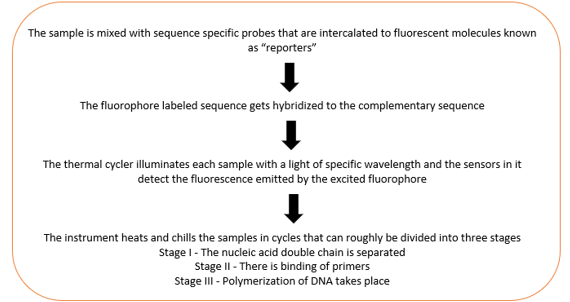
Quantitative Polymerase Chain Reaction (qPCR)
In RNA analysis, qPCR is a technique used to measure the number of expressed gene (overexpressed or under expressed) based on the amount of mRNA in the sample. The RNA sample should be reverse transcribed to complementary DNA (template) before this step. It is a modification of PCR and is carried out in a thermal cycler.15 Figure 2 depicts the various steps that occur in qPCR.
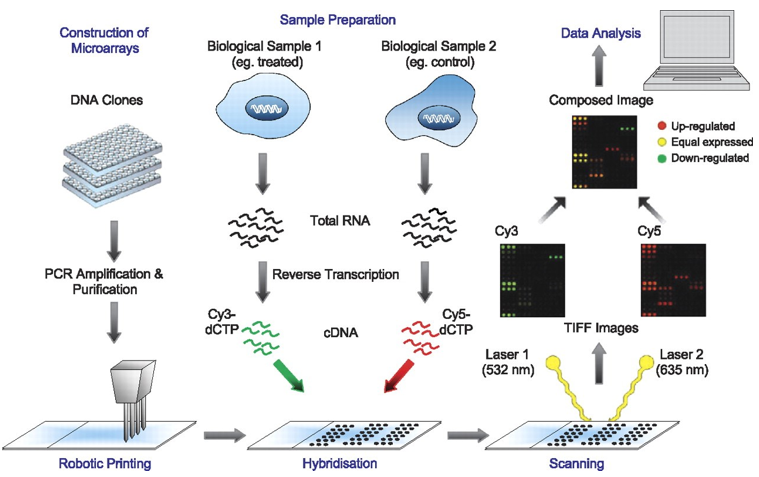
Microarray Technology
Microarray is a collection of several miniature spots of oligonucleotides located on a solid base. Each of such sequences is known as a probe and allows hybridization of cDNA or cRNA from a sample. The working principle behind microarray analysis is the hybridization between two strands of nucleic acids via hydrogen bonds. Microarray technology allows for the investigation of expression profile of a large set of genes.15 Figure 3 shows a pictorial representation about the basic steps performed during microarray technologies followed by qPCR.
DNA Hypermethylation
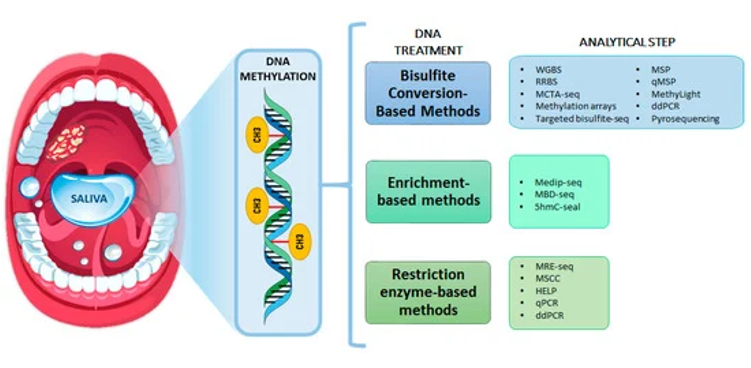
DNA methylation consists of the reversible addition of a methyl group to the carbon-5 position of the cytosine ring within CpG dinucleotides to form 5-methylcytosine (5-mC). The transfer is catalyzed by enzymes called as DNA methyltransferases. This process usually occurs in CpG islands and often located in gene promoter regions that are normally unmethylated.18
Different types of body fluids such as blood, sputum, bronchial lavage fluid, urine or saliva are used to detect aberrant DNA methylation. Saliva also represents a potential alternative to solid biopsy due to its noninvasive collection and its composition enriched with tumor biomarkers such as noncoding RNAs, proteins, mRNA, and genomic DNA. The detection of DNA methylation in saliva has emerged as a potential method for the early diagnosis of head and neck tumors.18
Viet et al carried out the very first study to evaluate promoter hypermethylation in salivary DNA from oral cancer patients in 2007. He and his associates analyzed the promoter hypermethylation of five genes (APC, ECAD, MGMT, p15, and p16) in oral cancer, oral dysplasia, and normal controls. Methylation of p16 was detected in 35% of oral cancer/dysplasia patients, MGMT and p15 methylation in 29%, APC in 14%, and ECAD in 7%.18
Guerrero-Preston et al performed a larger genome-wide DNA methylation study comprising of 27,578 CpG sites which revealed 301 potential tumor suppressor genes significantly hypermethylated in oral squamous cell carcinoma vs. normal tissues, 92 genes hypermethylated in leukoplakia vs. normal mucosa, and 143 hypermethylated genes in tumor vs. leukoplakia tissue.18
Over the last few years, several DNA-methylation studies in saliva have investigated the promoter hypermethylation status of various genes which had previously been identified as methylated in oral cancer tissue such as CDKN2A, MGMT, DAPK1, RASSF1A or ECAD.18
A study conducted by Nagata et al to analyze the promoter methylation of 13 genes in salivary oral rinses from 34 oral cancer patients and 24 healthy controls revealed that there were significantly higher levels of methylation for 8 genes (ECAD, MGMT, DAPK, RARb, p16, TMEFF2, WIF-1, and FHIT) in tumor salivary samples as compared to normal salivary samples. These results suggest the potential of salivary promoter DNA methylation for detecting oral cancer noninvasively.18 Figure 4 enlists the technologies for DNA-methylation evaluation.
Genetic Salivary Biomarkers with Potential for Oral Cancer Diagnosis
The genetic biomarkers are based either on changes in cellular DNA, such as promoter hypermethylation of p16 gene and Cyclin D1 gene amplification or altered mRNA transcripts such as presence of IL-8, IL-1 beta, OAZ1.19 Various genetic biomarkers that can be used to detect oral cancer as mentioned in Table 1 include:
p16
p16 is a protein that is known to slow down cell division by slowing down the progression of the cell cycle from the G1 phase to the S phase, thereby, acting as a tumor suppressor.20 Cancer cells show a significant increase in the accumulation of methylation in CpG islands in the promoter region of p16. These changes lead to a loss of the tumor suppressor gene function as methylation can physically inhibit the transcription of the gene, and methylation can also lead to the recruitment of transcription factors that repress transcription.21 Therefore, p16 can be used as a biomarker for the detection of oral cancer.
Cyclin D1
Cyclin D1 protein is encoded by CCND1 gene on long-arm of chromosome 11.22 CycD1 is contributes to oncogenesis by increasing anchorage-independent growth and angiogenesis through production of vascular endothelial growth factor (VEGF).23 Overexpression of CycD1 leads to down-regulation of Fas expression, which sequentially increases resistance to chemotherapy and protects from apoptosis.23 Overexpression of CycD1 is also correlated to reduced survival rates and increased metastasis.24
Antigen Ki-67
Ki-67 is encoded by gene MKI67 which is associated with cellular proliferation.25 Expression of Ki-67 is significantly elevated in malignant tissues, that have poor cellular differentiation, vis-à-vis normal tissues.26 Targeted cancer therapy towards Ki-97 have shown results as blocking of Ki-67 through microinjections of antibodies or with antisense oligonucleotides leads to arrest of cell proliferation.26
Table 1
Salivary biomarkersdetected by various biotechnological methods
|
Year of Publication |
Author |
Aim of Study |
Biotechnological Methodology |
Results |
Conclusion |
|
2004 |
Li et al 16 |
Salivary transcriptome diagnostics for oral cancer detection |
Qualitative Polymerase Chain Reaction (qPCR), microarray |
IL-8, IL-1β, DUSP1, HA3, OAZ1, S100P and SAT showed 3.5-fold elevation in saliva of OSCC patients |
Salivary transcriptomes show a promising avenue as a diagnostic biomarker for OSCC |
|
2009 |
Park et al 27 |
Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection |
Reverse Transcription- preamplification- qualitative Polymerase Chain Reaction (RT-preamp-qPCR) |
Salivary levels of miRNA-200a and miRNA-125a are lower in patients with OSCC compared to healthy controls. |
Salivary miRNA can use a diagnostic tool for OSCC due to differential expression compared to healthy individuals |
|
2010 |
Markopoulos et al 19 |
Salivary markers for oral cancer detection |
N/A |
Various biomarkers such as IL-8, IL-1β, CycD1 and Ki-67 have been detected at higher concentrations in saliva of patients with OSCC |
OSCC can be diagnosed by testing salivary molecular markers in samples from subjects |
|
2011 |
Shah et al 28 |
A review on salivary genomics and proteomics biomarkers in oral cancer |
N/A |
CycD1, Ki67- Diagnosis, prognosis & post operative monitoring. |
Salivary genomic biomarkers aid in effective screening to identify patients with high risk of OSCC as well as designing better treatment modalities |
|
2012 |
Liu et al 29 |
Exploiting salivary miR-31 as a clinical biomarker of oral squamous cell carcinoma |
Quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) |
Salivary levels of miR-31 were significantly increased in OSCC patients, in all stages. miR-31 was more abundant in saliva than plasma. Levels of miR-31 showed reduction after excision of tumor. |
Salivary levels of miR-31 are sensitive marker for OSCC due to its exclusive appearance in OSCC compared to oral leukoplakia and is primarily sourced for the tumor site. |
|
2012 |
Elashoff et al 30 |
Pre-validation of salivary biomarkers for oral cancer detection |
Real Time Polymerase Chain Reaction (PCR) |
Expression of all 7 mRNA and 3 protein markers increase in OSCC in all 5 cohorts compared to the controls |
Association of biomarkers highly consistent with OSCC, especially IL-8, SAT |
|
2013 |
Tang et al 31 |
Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis |
Reverse Transcription- qualitative Polymerase Chain Reaction (RT-qPCR) |
Long, non-coding RNA HOTAIR was detected significantly in saliva of patients with lymph node metastasis |
ln RNA HOTAIR might be an appropriate diagnostic biomarker for OSCC metastasis. |
|
2014 |
Cheng et al 32 |
Levels of potential Oral Cancer salivary mRNA biomarkers in Oral Cancer patients in remission and Oral Lichen Planus patients |
Pre-amplification RT-qPCR approach with nested gene-specific primers |
Salivary levels of OAZ1, S100P, DUSP1- significantly higher in newly diagnosed OSCC patientsNo significant difference in levels of IL-8, IL-1β, H3F3A & SAT1 |
Salivary OAZ1, S100P & DUSP1- candidates for diagnosis of OSCC in remission & in OLP patients |
Table 0
|
Table 1 Cont.. |
|||||
|---|---|---|---|---|---|
|
2017 |
Cheng et al33 |
Chronic periodontitis can affect the levels of potential oral cancer salivary mRNA biomarkers |
Pre-amplification reverse transcription-quantitative Polymerase Chain Reaction (RT-qPCR) with nested gene-specific primers |
Only S100P levels increase in OSCC compared to chronic periodontitis patient (smoking or non-smoking) No significant difference for salivary levels of IL-8, IL-1β, DUSP1 with non-smoking patients, and no difference reported for OAZ1 and SAT for smoking patients |
Salivary S100P mRNA is a reliable biomarker for detection of OSCC, regardless of the presence of Chronic Periodontitis |
|
2017 |
Menke et al34 |
More accurate oral cancer screening with fewer salivary biomarkers |
N/A |
Five most sensitive biomarkers for OSCC were- CALCA, RMB6, S100A2, HOXA9 and most specific were - RASSF1A, AIM1, ESR, p16, DCC |
Elevated levels of ESR can be used to rule out presence of cancer with a negative report. |
|
2019 |
Liu et al35 |
Salivary LDOC1 is a gender-difference biomarker of oral squamous cell carcinoma |
Quantitative Reverse Transcription- Polymerase Chain Reaction (qRT-PCR) |
Salivary levels of LDOC1 were upregulated in females with in females with OSCC (p=0.0072) and significantly downregulated in males with OSCC (p=0.0206) |
High levels of salivary LDOC1 expression can act as a diagnostic tool for OSCC for females. |
|
2020 |
Rivera et al36 |
Biomarkers of progression to oral cancer in patients with dysplasia |
N/A |
ALDH1A1, PROM1 - correlated with malignant transformation in patients with premalignant leukoplakia PDPN- valuable for risk assessment of malignant transformation in patients with oral leukoplakia |
Potential biomarkers indicating for malignant transformation and for the absence of signs of disease after treatment |
|
2020 |
Oh et al37 |
Potential salivary mRNA biomarkers for early detection of oral cancer |
Microarray, real time qPCR |
Six salivary mRNA- MAOB, NAB2, COL3A1, CYP27A1, NPIPB4, SIA3 were significantly decreased in the OSCC group compared to the control |
Two mRNA combinations of CYP27A1+SIA3 and MAOB+NAB2 showed the most promise as potential diagnostic biomarkers. |
|
2021 |
Romani et al38 |
Genome-wide study of salivary miRNAs identifies miR-423-5p as promising diagnostic and prognostic biomarker in oral squamous cell carcinoma |
Microarray, quantitative real time PCR |
Twenty-five miRNAs found to expressed differently in OSCC patients. Seven of them were significantly associated with disease-free survival (DFS). miR-106b-5p, miR-423-5p and miR-193b-3p have the best diagnostic performance. |
miR-160b-5p, miR-423-5p and miR-193b-3p are expressed at high levels in OSCC patients and combination provides the best diagnostic performance. |
|
2021 |
Ueda et al39 |
Salivary CPLANE1 Levels as a Biomarker of Oral Squamous Cell Carcinoma |
Quantitative Reverse Transcription PCR (qRT-PCR) |
Salivary levels of CPLANE1 were higher in patients with OSCC compared to healthy volunteers and OMPDs patients. |
Salivary CPLANE1 can act as useful diagnostic tool for OSCC screening and early detection. |
|
2021 |
Rapado-Gonzáles et al18 |
Saliva Gene Promoter hypermethylation as a biomarker in oral cancer |
N/A |
Methylation of various gene promoters such as p16, MGMT, RASSF1A, TIMP3, N1D2, TRH found in different types of oro-, head and neck, and oropharyngeal cancers |
Salivary DNA methylation is a suitable diagnostic tool for oral cancer diagnosis and prognosis |
|
2021 |
Osan et al40 |
The connection between MicroRNAs and Oral Cancer pathogenesis: emerging biomarkers in oral cancer management |
N/A |
MicroRNAs miR-21, MiR-184, miR-211, miR-31 such as are associated with dysregulation of various enzymes and proteins in the process of oncogenesis. |
MicroRNAs, particularly miR-31 are a possible diagnostic tool for oral cancer diagnosis |
|
2022 |
Rapado-Gonzáles et al41 |
Integrity and quantity of salivary cell-free DNA as a potential molecular biomarker in oral cancer: A preliminary study |
Fluorometry method, quantitative real-time polymerase chain reaction (qPCR) |
Salivary cfDNA integrity indexes: ALU115/ALU60 and ALU247/ALU60 were significantly higher in OSCC patients. |
Salivary cfDNA integrity indexes (ALU115/ALU60 and ALU247/ALU60) show potential to be non-invasive diagnostic biomarkers for OSCC. |
Interleukins: IL-8, IL-1Β
Interleukin-8, though a chemokine of the CXC family, is responsible as an activator and chemoattractant for neutrophils.42 It plays a role in proliferation of tumor angiogenesis, cell adhesion and cell cycle arrest.13 According to Khurshid et al, it is vital for chemotaxis of macrophages and granulocytes that are seen in the stroma of OSCC predominantly.10 Interleukin-1 beta (IL-1β) is another cytokine that shows elevated salivary levels in OSCC. Its role in carcinogenesis is that of a chemical mediator for cell proliferation, differentiation, and apoptosis.13 The detection of both is performed using ELISA.16
Ornithine decarboxylase antizyme 1 (OAZ1)
Ornithine decarboxylase antizyme (OAZ) 1 function as an inhibitor to ornithine decarboxylase (ODC), which is a rate-limiting enzyme in polyamine synthesis.43 Based on its inhibitory function, OAZ1 is suggested to work as a tumor suppressor.16 Based on their observations of elevated salivary levels of OAZ1 mRNA, Cheng et al suggest that there is presence of increased polyamine level in OSCC. 32
S100 calcium binding protein P (S100P)
The protein encoded by this gene is belongs to the S100 family of proteins that contain 2 EF-hand calcium-binding motifs.44 It is responsible for cell cycle regulation and differentiation during carcinogenesis.45 Surge in S100P mRNA expression is seen in anoikis-resistant OSCC cells in contrast to the anoikis-sensitive cells, which demonstrates S100P involvement in OSCC metastasis.46 Further evidence is required for validation as a biomarker.32
Ras association domain-containing protein 1 A (RASSF1A)
RASSF1A is one of the eight isoforms of RASSF1 gene.47 Though lacking apparent enzymatic activity, RASSF1A contains a Ras association (RA) and thus a potential effector of the Ras oncoprotein. RASSF1A is responsible for modulating multiple apoptotic and cell cycle checkpoint pathways. The most common reason for the loss or reduction in function of RASSF1A is transcriptional silencing by inappropriate promoter methylation. Subsequentially, numerous tumor suppressors can be inactivated by this epigenetic mechanism, leading to oncogenesis.48 Thus, RASSF1A promoter hypermethylation provides an epigenetic tumor biomarker for diagnosis.47
MicroRNA (miRNA)
miRNA is a small single stranded non-coding RNA fragment, that has about 9-25 base pairs.48 They are secreted via exosomes and by repressing or activating translocation in local or distant cells, they coordinate cellular and tissue programming.48 Dysregulation in expression of miRNA would consequentially affect cell growth and would act as tumor suppressors or oncogenes in various cancers.7 As their expression increases to significant proportion- about 10-100 fold increase, compared to messenger RNA, they can be used as potential tumor biomarkers.5 Some potential miRNA biomarkers include: miRNA-125a (has effect on genes in MAPK pathway and in cell proliferation), miRNA-200a (essential role in tumor suppression and early metastasis) and miRNA-31 (acts as a tumor suppressor).5
Cell-free DNA (cfDNA)
CfDNA is a type of DNA that is released into the blood originating from either a healthy cell, tumor cell or cells from the tumor micro environment. Cause for release is due to cellular processes such as oncosis, netosis, apoptosis, necrosis.7 CfDNA predominantly circulates as a nuclear complex of histones and DNA: nucleosomes.49 The various methods for detection of cfDNA include are by PCR-based sequence specific detection and DNA sequencing in form of general genomic analysis of all the cfDNA that is found in the blood.50
Limitations of Salivary Biomarkers for Cancer Detection
The field of using salivary tumor markers for various diagnostic and prognostic purposes has been under consideration the past two decades. Despite the discovery of multiple markers with various degrees of sensitivity and specificity, no one marker has been established as the ‘gold standard’ diagnostic criteria for the diagnosis of oral squamous cell carcinoma. The need of the future research is to establish a standardized method for collection of the sample to ensure uniform processing and analysis of the sample. The biomarkers need to be validated under various inflammatory and other cancerous lesions to establish a highly specific biomarker for OSCC diagnosis.
Conclusion
This systematic review revealed that genomic salivary biomarkers such as p16, Cyclin D1, OAZ1, Antigen Ki-67, IL-8 and IL-1β, S100P and RASSF1A have been detected in the salivary samples of cancer patients. Different molecular technologies such as PCR, qPCR, microarray technologies and DNA hypermethylation have been employed to detect their expression. But their role is still limited as an adjunct to biopsy which still stands as the ‘gold standard’. Further studies need to be conducted before salivary biomarkers alone can aid in the diagnosis of oral cancer.